Code
library(Data4Ecologists) # for data
data(Selake)Be able to identify when to use a mixed model
Learn how to implement mixed models in R/JAGs when the response is Normally distributed (linear mixed effect models, lme)
Be able to describe models and their assumptions using equations and text and match parameters in these equations to estimates in computer output.
Next chapter:
Selenium, Se, a bi-product of burning coal is measured in…
Goal: determine the relationship between mean (log) Se in lake and mean (log) Se in fish.
What are the consequences of ignoring the fact that we have multiple observations from each lake?
What strategies might we use to analyze these data?
Note: our main question involves a predictor-response relationship in which the predictor is constant within each cluster or sample unit
Strategies:
Lets do this!
When you have more than one measurement on the same observational unit
Experiments or surveys with multiple sizes of sample units
When you want to generalize to a larger population of sample units
Key features:
Why are they so popular:
Study objective: investigate tradeoffs between growth rate, size, and lifespan of Mountain pines (Pinus montana) in Switzerland (Bigler, 2016).
Is it better to grow quick but die young? Grow more slowly and live longer?
Sample units:
Variables:
Sampling Effort:
Interest lies in modeling:
Using macro-fauna and abiotic variables:
Linear regression assumes that observations are independent. Is that reasonable in this case?
Think of models at 2 levels:
Stage 1 (level 1 model):
Stage 2 (level 2 model):
Can be useful exploratory approach when you have lots of data for each cluster, but few clusters
NAP is a “level-1” covariate (it varies within each cluster)
exposure is a “level-2” covariate (it is constant within a cluster)
Let \(R_{ij}\) = the species richness for the \(j^{th}\) sample on the \(i^{th}\) beach (note: we now need two subscripts!)
Level 1 model: model for observations within each cluster (i.e., for each beach)
\(R_{ij} = \beta_{0i} + \beta_{1i}NAP_{ij} + \epsilon_{ij}\); (\(j = 1, 2, \ldots, 5\) observations for each Beach)
Each beach has its own intercept \(\beta_{0i}\) and slope \(\beta_{1i}\)
RIKZdat$NAPc = RIKZdat$NAP-mean(RIKZdat$NAP) #center NAP variable
Beta<-matrix(NA, 9,2) # to hold slope and intercepts
Exposure<-matrix(NA,9,1) # to hold exposure level for each beach
for(i in 1:9){
Mi<-lm(Richness~NAPc, data=subset(RIKZdat, Beach==i))
Beta[i,]<-coef(Mi)
Exposure[i]<-subset(RIKZdat, Beach==i)$exposure.c[1]
}
betadat <- data.frame(Beach = 1:9, intercept = Beta[,1],
slope = Beta[,2], exposure.c = Exposure)Note: I have centered the NAP variable
NAPThis gives us a data frame of coefficients and level-2 predictors for a level-2 model:
Beach intercept slope exposure.c
1 1 10.692614 -0.3718279 Low
2 2 11.893999 -4.1752712 Low
3 3 2.790385 -1.7553529 High
4 4 2.653600 -1.2485766 High
5 5 9.688335 -8.9001779 Low
6 6 3.841864 -1.3885120 High
7 7 2.992969 -1.5176126 High
8 8 4.293257 -1.8930665 Low
9 9 5.263276 -2.9675304 LowFor a tidyverse solution - see book/R code.
Model for the slope and intercept parameters (analyze the summary statistics, \(\hat{\beta}_{0i}, \hat{\beta}_{1i})\) using level-2 predictors (ones that are constant within a cluster)
For now, ignore the fact that the variability of \(b_{0i}, b_{1i}\) seems to depend on exposure level (“low”, “high”).
Call:
lm(formula = intercept ~ exposure.c, data = betadat)
Residuals:
Min 1Q Median 3Q Max
-4.0730 -0.4161 -0.0767 1.3220 3.5277
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.070 1.291 2.378 0.0491 *
exposure.cLow 5.297 1.732 3.058 0.0184 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.582 on 7 degrees of freedom
Multiple R-squared: 0.5719, Adjusted R-squared: 0.5107
F-statistic: 9.349 on 1 and 7 DF, p-value: 0.01838
Call:
lm(formula = slope ~ exposure.c, data = betadat)
Residuals:
Min 1Q Median 3Q Max
-5.2386 -0.2778 0.0890 0.6940 3.2897
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.478 1.229 -1.202 0.268
exposure.cLow -2.184 1.649 -1.325 0.227
Residual standard error: 2.458 on 7 degrees of freedom
Multiple R-squared: 0.2005, Adjusted R-squared: 0.08625
F-statistic: 1.755 on 1 and 7 DF, p-value: 0.2268Level-1 Model:
Level-2 Model:
Substitute into level-1 equation to get the composite equation
\[R_{ij} = (\beta_0 + \gamma_{0}Exposure_i+b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \epsilon_{ij}\]
\[R_{ij} = (\beta_0 + b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \gamma_0Exposure_{i} + \epsilon_{ij}\]
\(\Rightarrow\) random intercepts and slopes model (or random coefficients model)
Rather than use a 2-stage approach, we could just posit a model for the data using random and fixed effects.
Random Intercepts Model:
\[R_{ij} = \beta_0 + b_{0i}+ \beta_1NAP_{ij} + \beta_2Exposure_i + \epsilon_{ij}\] \[b_{0i} \sim N(0,\sigma^2_{b_{0}})\] \[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
Random Intercepts and Slopes Model:
\[R_{ij} = (\beta_0 + b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \beta_2Exposure_{i} + \epsilon_{ij}\] \[(b_{0i}, b_{1i}) \sim N(0,D)\] \[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
Random Intercepts and Slopes Model:
\[R_{ij} = (\beta_0 + b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \beta_2Exposure_{i} + \epsilon_{ij}\] \[(b_{0i}, b_{1i}) \sim N(0,D)\] \[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
We can think of \(b_{0i}\) and \(b_{1i}\) as deviations from the average intercept (\(\beta_0\)) and slope (\(\beta_1\)), respectively.
Or, we can think in terms of beach-level intercepts and slopes:
\[R_{ij} = (\beta_{0i}) + (\beta_{1i})NAP_{ij} + \beta_2Exposure_{i} + \epsilon_{ij}\]
Two popular packages: nlme and lme4:
nlme (older)
glmmTMB, INLA);lme4 (newer)
year and Beach if we had many years of data).Many others too…see glmm wikidot
Two others that we will consider:
For now, let’s fit the random intercept and random intercept and slope models using the lmer function in the lme4 package!
Random Intercepts Model:
\[R_{ij} = \beta_0 + b_{0i}+ \beta_1NAP_{ij} + \beta_2Exposure_i + \epsilon_{ij}\] \[b_{0i} \sim N(0, \sigma^2_{b_{0}})\] \[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
Fit this model in R using lmer and identify \(\hat{\beta}_0\), \(\hat{\beta}_1\), \(\hat{\sigma}^2_\epsilon\), \(\hat{\sigma}^2_{b_{0}}\)!
Random Intercepts and Slopes Model:
\[R_{ij} = (\beta_0 + b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \beta_2Exposure_{i} + \epsilon_{ij}\] \[(b_{0i}, b_{1i}) \sim N(0,D)\]
\[D = \begin{bmatrix} var(b_{0i}) & cov(b_{0i},b_{1i}) \\ cov(b_{0i},b_{1i}) & var(b_{1,i}) \end{bmatrix}\]
\[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
Fit this model in R and identify the different parameters:
We can write our models in two ways…
\[R_{ij} = (\beta_0 + b_{0i}) + \beta_1NAP_{ij} + \beta_2Exposure_{i} + \epsilon_{ij}\] \[b_{0i} \sim N(0,\sigma^2_{b_{0}})\] \[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
\[R_{ij} | b_{0i} \sim N(\mu_i, \sigma^2_\epsilon)\] \[\mu_i = (\beta_0 + b_{0i}) + \beta_1NAP_{ij} + \beta_2Exposure_{i}\] \[b_{0i} \sim N(0,\sigma^2_{b_{0}})\]
With either approach, we have 2 random processes that are both formulated using a Normal distribution.
The latter will more easily translate to other statistical distributions (generalized linear mixed effects models).
Later, we will see that for this model, we can derive the marginal distribution of \(R_{ij}\) and refer to it rather than \(R_{ij} \mid b_{0i}\)
If you are a Bayesian, you can ignore the distinction between “prediction” and “estimation”…ALL parameters are random variables!
Demonstration w/ R code.
Each beach also has its own intercept. What if we modeled Beach using fixed effects? . . .
Call:
lm(formula = Richness ~ factor(Beach) - 1 + NAPc, data = RIKZdat)
Residuals:
Min 1Q Median 3Q Max
-4.8518 -1.5188 -0.1376 0.7905 11.8384
Coefficients:
Estimate Std. Error t value Pr(>|t|)
factor(Beach)1 8.9392 1.4301 6.251 3.61e-07 ***
factor(Beach)2 12.0173 1.3690 8.778 2.29e-10 ***
factor(Beach)3 2.5343 1.3796 1.837 0.074716 .
factor(Beach)4 2.9063 1.3723 2.118 0.041364 *
factor(Beach)5 8.0409 1.3746 5.850 1.22e-06 ***
factor(Beach)6 3.7161 1.3697 2.713 0.010271 *
factor(Beach)7 3.5025 1.3934 2.514 0.016705 *
factor(Beach)8 4.3862 1.3707 3.200 0.002920 **
factor(Beach)9 5.1572 1.3731 3.756 0.000629 ***
NAPc -2.4928 0.5023 -4.963 1.79e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.06 on 35 degrees of freedom
Multiple R-squared: 0.8719, Adjusted R-squared: 0.8353
F-statistic: 23.82 on 10 and 35 DF, p-value: 9.56e-13Fixed effects:
Random effects:
exposure.c since it is constant for each Beach (and therefore, confounded with the Beach coefficients)factor(Beach)1 factor(Beach)2 factor(Beach)3 factor(Beach)4 factor(Beach)5
8.939200 12.017303 2.534266 2.906323 8.040936
factor(Beach)6 factor(Beach)7 factor(Beach)8 factor(Beach)9 NAPc
3.716094 3.502535 4.386168 5.157177 -2.492836
exposure.cLow
NA Beach and NAP (another 8 parameters)Shrinkage depends on:
\[R_{ij}|b_i \sim N(\mu_i, \sigma^2_\epsilon)\] \[\mu_i = (\beta_0 + b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \beta_2Exposure_{i}\] \[b_i = (b_{0i}, b_{1i}) \sim N(0, D) \text{ with}\] \[D = \begin{bmatrix} var(b_{0i}) & cov(b_{0i},b_{1i}) \\ cov(b_{0i},b_{1i}) & var(b_{1,i}) \end{bmatrix}\]
Subject-Specific (lines for a particular beach):
Subject-Specific (line for a “typical” beach with \(b_{0i} = b_{1i} = 0)\):
Population Average (averages over beaches):
R for a demonstration!
Random Intercepts and Slopes Model:
\[R_{ij} = (\beta_0 + b_{0i}) + (\beta_1+b_{1i})NAP_{ij} + \beta_2Exposure_{i} + \epsilon_{ij}\] \[(b_{0i}, b_{1i}) \sim N(0,D)\] \[\epsilon_{ij} \sim N(0, \sigma^2_\epsilon)\]
What are our assumptions?
Plot of within beach residuals, \(\hat{\epsilon_{ij}}\), versus beach-level predictions \({\hat{R}_{ij}} = \hat{\beta}_0 + \hat{b}_{0i} + (\hat{\beta}_1+\hat{b}_{1i})NAP + \hat{\beta}_2(exposure=low)\)
check_model function offers many more checks
See R for a demonstration!
library(nlme)
lme.fit<-lme(Richness~NAPc+exposure.c, random=~1|Beach, data=RIKZdat)
summary(lme.fit)Linear mixed-effects model fit by REML
Data: RIKZdat
AIC BIC logLik
240.5538 249.2422 -115.2769
Random effects:
Formula: ~1 | Beach
(Intercept) Residual
StdDev: 1.907175 3.059089
Fixed effects: Richness ~ NAPc + exposure.c
Value Std.Error DF t-value p-value
(Intercept) 3.170680 1.1739987 35 2.700752 0.0106
NAPc -2.581708 0.4883901 35 -5.286160 0.0000
exposure.cLow 4.532777 1.5755610 7 2.876929 0.0238
Correlation:
(Intr) NAPc
NAPc -0.028
exposure.cLow -0.746 0.037
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-1.5163203 -0.4815106 -0.1218700 0.2922854 3.8777562
Number of Observations: 45
Number of Groups: 9 Degrees of Freedom (differ for level-1 and level-2 predictors):
NAPc = 35exposure.cLow = 7Level-1: within-subjects degrees of freedom calculated as the number of observations minus the number of groups minus the number of level-1 regressors in the model.
The formula are not important….what is:
NAP on species richness than exposure since NAP varies between and within beaches.lme accounts for the data structure when carrying out statistical tests.Note: lme’s df are essentially correct for balanced data (all clusters have an equal number of observations). For unbalanced data, the tests (and df) are only approximate.
lmer in lme4lmerTest package and Section 18.12.3 of the book and R code).AIC comparisons and likelihood ratio tests are complicated by the fact that the variance parameter is “on the boundary”
See Bolker’s FAQ
Number of parameters for calculating AIC also depends on focus (on individual subjects or population)
See Bolker’s FAQ
See LectureMixedMods.Rmd for an option, or have a look at the RLRsim or pbkrtest packages for simulation-based alternatives.
For nested models, generate a null distribution for likelihood ratio test statistic = - 2(LogL(model1)-LogL(model2)).
p-value = proportion of simulated observations that are as extreme, or more extreme than the likelihood ratio statistic calculated using the observed data.
REML = Restricted Maximum Likelihood (usual default method)
General Recommendation
Fixed and random effects can “compete” to explain patterns in your response variable… . . .
lm(y ~ x1)lmer(y ~ 1 + (1 | site))lmer(y ~ x1 + (1 | site))lmer(y ~ x1+(1+x1|site))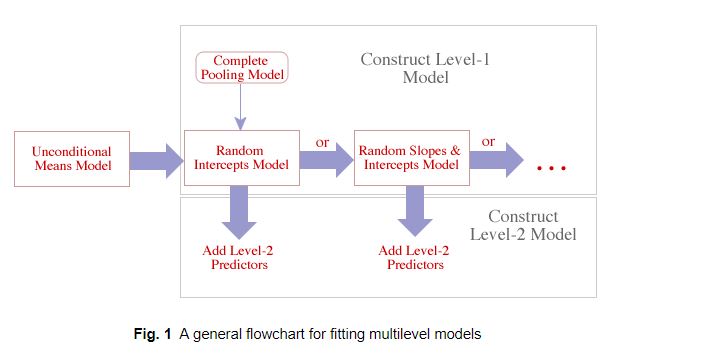
Schematic from Jack Weiss’s course site.
Pick the best of these, then add level-2 predictors (predictors that are constant within clusters).
Strategy outlined by:
Singer, J. D. and Willett, J. B. (2003) Applied Longitudinal Data Analysis: Modeling Change and Event Occurrence. (Oxford University Press, Oxford, UK).
Although random intercepts models are common. . .
Schielzeth and Forstmeier (2009) suggest random slopes are usually appropriate for level-1 predictors (i.e., when x varies within a subject).
Attempt to make inference from a maximal model:
Lots of debate on how best to approach model building/selection.
\[R_{ij} = \beta_0 + b_{0i}+ \beta_1NAP_{ij} + \beta_2Exposure_i + \epsilon_{ij}\]
Variance of \(R_{ij} = var(b_{0i}+\epsilon_{ij}) = var(b_{0i}) +var(\epsilon_{ij}) = \sigma^2_{b_{0}} + \sigma^2_\epsilon\)
Covariance (\(Y_{ij}, Y_{ij^\prime}) = \sigma^2_{b_{0}}\) (2 observations, same cluster [beach] since they share \(b_{0i}\))
Covariance (\(Y_{ij}, Y_{i^\prime j}) = 0\) (2 observations taken from 2 different clusters [beaches])
Intraclass correlation = Cor\((Y_{ij}, Y_{ij^\prime})\) = \(\frac{\sigma^2_{b_{0}}}{\sigma^2_{b_{0}}+\sigma^2_\epsilon} = 0.28\), correlation among observations taken from the same cluster.
Different levels of correlation induced by (1 | PACKID) + (1 | Year)
R for demonstration!
\[Y_{i} = X_{i}\beta + Z_ib + \epsilon_{i}\] \[\epsilon_i \sim N(0, \Sigma_i)\] \[b \sim N(0, D)\]
\[Y_i|b \sim N(X_i\beta+Z_ib, \Sigma_i)\] \[b \sim N(0, D)\]
If we average over (or integrate out) the random effects, we get the marginal Distribution of \(Y\).
\[Y_i \sim N(X_i\beta, V_i), V_i = Z_iDZ_i^\prime + \Sigma_i\]
This is the likelihood that R maximizes to get \(\hat{\beta}, \hat{D}, \hat{\Sigma_i}\).
For random intercepts model:
\[Y_{i}\sim N(X_i\beta, V_i)\]
\(V_i = \begin{bmatrix} \sigma^2 & \rho & \cdots & \rho \\ \rho & \ddots & \ddots & \vdots \\ \vdots & \ddots & \ddots & \rho \\ \rho & \cdots & \rho & \sigma^2 \end{bmatrix}\) \(\rho = \frac{\sigma^2_{b_{0}}}{\sigma^2_{b_{0}}+\sigma^2_\epsilon}\)
Var/Cov matrix for \(Y\) (all data) = \(\begin{bmatrix} V_i & 0 & \cdots & 0 \\ 0 & \ddots & \ddots & \vdots \\ \vdots & \ddots & \ddots & 0 \\ 0 & \cdots & 0 & V_i \end{bmatrix}\)
glsWe might have posited this model directly.
We can fit it using the gls function in the nlme library
The gls function also allows for:
See Ch 4 Zuur et al. and the section of the course on gls models.
glsgls.fit<-gls(Richness~NAPc+exposure.c, method="REML",
correlation=corCompSymm(form=~1|Beach),
data=RIKZdat)
summary(gls.fit)Generalized least squares fit by REML
Model: Richness ~ NAPc + exposure.c
Data: RIKZdat
AIC BIC logLik
240.5538 249.2422 -115.2769
Correlation Structure: Compound symmetry
Formula: ~1 | Beach
Parameter estimate(s):
Rho
0.2798938
Coefficients:
Value Std.Error t-value p-value
(Intercept) 3.170680 1.1739987 2.700752 0.0099
NAPc -2.581708 0.4883901 -5.286160 0.0000
exposure.cLow 4.532777 1.5755610 2.876929 0.0063
Correlation:
(Intr) NAPc
NAPc -0.028
exposure.cLow -0.746 0.037
Standardized residuals:
Min Q1 Med Q3 Max
-1.5551728 -0.6415409 -0.1554932 0.4150315 3.3566242
Residual standard error: 3.604905
Degrees of freedom: 45 total; 42 residual